Order Statistics and Statistics of Extremes¶
Supramolecular assemblies of biological interest, such as multimeric proteins, protein oligomers and fibers, all have modular architecture (see Figure). For example, fibrin protofibrils consist of fibrin monomers polymerized both in the longitudinal and in the lateral direction. Dynamic force measurements carried out on these systems show that domain-domain interactions modulate the physical properties of the individual protein repeats, which results in their strong coupling. We developed a new theory, inspired by Order Statistics, which captures the time-ordered nature of microscopic transitions in the biological matter. For example, the mechanically driven unfolding transitions in a generic hexamer \((D)_6\), subject to a force-clamp, which occurs in a time-ordered fasion, i.e. \(t_{1:6} \leq t_{2:6} \leq \cdots \leq t_{6:6}\), are described by the Order Statistics cumulative distribution function (cdf) \(\Phi_{i:6}(t)\), and the probability density function (pdf) \(\phi_{i:6} = d\Phi_{i:6}(t)/dt\) (\(i=1, 2, \cdots, 6\)). On the other hand, when the force-driven unfolding transitions in \((D)_6\) are induced by an applied force-ramp, the recorded peak forces increase in the sequence \(f_{1:6} \leq f_{1:5} \leq \cdots \leq f_{1:1}\), where \(f_{1:n}\) (\(n=6, 5, \cdots, 1\)) are the unfolding forces. Here, the experimentally measurable quantities are the cdf, \(\Phi_{i:6}(t)\), and the pdf, \(\phi_{i:6} = d\Phi_{i:6}(t)/dt\) (\(i=1, 2, \cdots ,6\)). We attemp to develop the Statistical Mechanics approach based on Order Statistics for correlated random variables to describe the mechanical unfolding reaction in protein oligomers and fibers, in order to probe protein-protein interactions and to characterize domain stabilization effects.
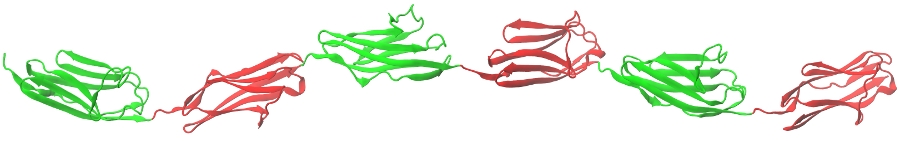
Figure: A ribbon representation of the oligomer \((D)_6\), formed by head-to-tail connected protein domains of the human immunoglobulin (Ig) from human titin.
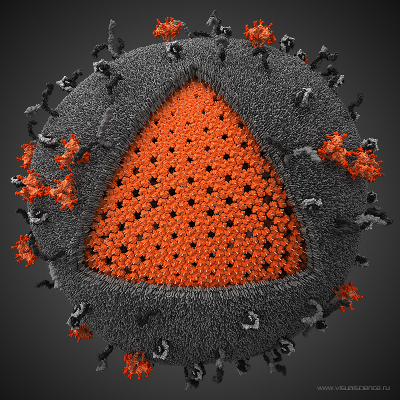
Figure: Human Immunodeficiency Virus - HIV (picture is taken from Visual Science).
Biologically important supramolecular assemblies, such as viral capsids of many animal and plant viruses, have modular architecture of different geometries (see Figure). For example, virus shells are formed by self-assembled identical units called capsomers, which interact together to form the overall 3D structure. Atomic force measurements carried out on these systems show that domain-domain (inter-capsomer) interactions determine the structural stability and the functional integrity of viral shells. We develop a new theory, which is based on Statistics of extreme observations (Statistics of Extremes) to characterize the physical properties of viral shells. For example, structural damage to a viral capsid occurs when any one of \(N\) edges between a pair of adjacent capsomers splits apart first at the minimum time \(t_{1:N}\). The force-induced unfolding transition in a protein domain and the unbinding transition in a ligand-repector complex can also viewed as an extreme outcome in a long sequence of many (unfolding and unbinding) attempts. For example, an unfolding (unbinding) transition in a protein (ligand-receptor complex) corresponds to the last \(N\)-th attempt (out of \(N\) attempts) to overcome the free energy barrier for unfolding (unbinding) at the maximum (longest) time \(t_{N:N}\). The minima and maxima, which correspond to the extreme variates \(x_{1:N}\) and \(x_{N:N}\), where \(x\) is a dynamic observable, are studied by Statistics of Extremes. Because equilibrium information is contained in the tail of the distributions, this theory can be used to resolve the equilibrium free energy landscape for the microscopic transitions. We develop Statistics of Extremes for correlated random variables to model the dissociation of biomolecular complexes and aggregates, and to explore the mechanical limits of viral shells.