Microtubule biomechanics and tubulin bond energies¶
In a close collaboration with experimental groups of prof. E. L. Grishchuk (University of Pennsylvania, School of Medicine) and prof. F. I. Ataullakhanov (National Hematology Research Center, Russia) we work on exploring physico-chemical properties of microtubules, as well as their essencial role in mitosis and cell division.
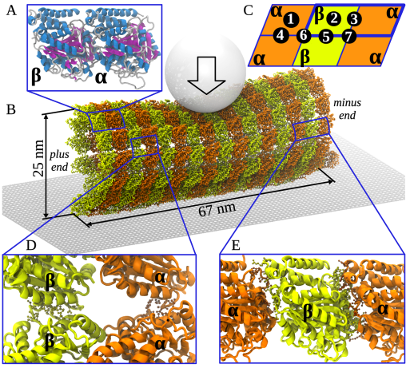
Microtubules (MT), the primary components of the chromosome segregation machinery, are stabilized by longitudinal and lateral non-covalent bonds between the tubulin subunits. However, the thermodynamics of these bonds and the microtubule physico-chemical properties are poorly understood. We applied SOP-GPU package to explore the biomechanics of microtubule polymers using multiscale computational modeling and nanoindentations in silico of a contiguous microtubule fragment. A close match between the simulated and experimental force-deformation spectra enabled us to correlate the microtubule biomechanics with dynamic structural transitions at the nanoscale.
Our mechanical testing revealed that the compressed MT behaves as a system of rigid elements interconnected through a network of lateral and longitudinal elastic bonds. The initial regime of continuous elastic deformation of the microtubule is followed by the transition regime, during which the microtubule lattice undergoes discrete structural changes, which include first the reversible dissociation of lateral bonds followed by irreversible dissociation of the longitudinal bonds. We have determined the free energies of dissociation of the lateral (\(6.9 \pm 0.4\) kcal/mol) and longitudinal (\(14.9 \pm 1.5\) kcal/mol) tubulin-tubulin bonds. These values in conjunction with the large flexural rigidity of tubulin protofilaments obtained (\(18,000-26,000\) pN nm \(^2\)), support the idea that the disassembling microtubule is capable of generating a large mechanical force to move chromosomes during cell division. Our computational modeling offers a comprehensive quantitative platform to link molecular tubulin characteristics with the physiological behavior of microtubules. The developed in silico nanoindentation method provides a powerful tool for the exploration of biomechanical properties of other cytoskeletal and multiprotein assemblies.
The results was published in J. Am. Chem. Soc. (2014).