Dynamic behavior of plant & animal viral capsids¶
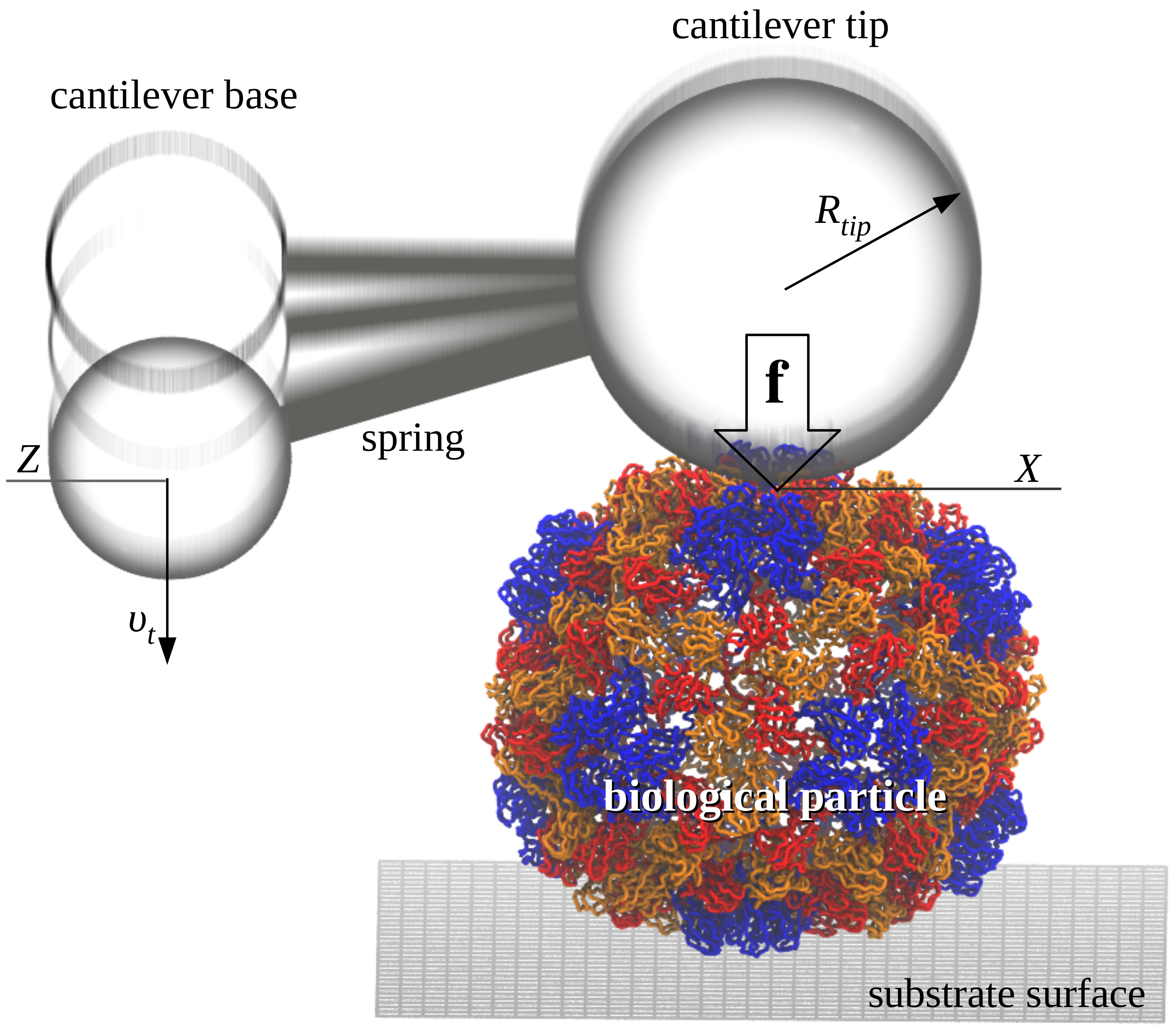
The SOP-GPU package is being used by graduate students in our research group to carry out dynamic force experiments in silico on the viral capsids CCMV, HK97, and \(\phi 29\). The results of molecular simulations of the mechanical indentation on these biological assemblies are compared directly with the experimental force-indentation curves from our collaborator Dr. Wuite lab (University of Amsterdam). We seek to characterize the physical properties of viral shells as a function of their geometry and other factors (pH and site mutations), under the experimentally relevant conditions of force application, including the force-loading rate, and the spring constant and size of a cantilever tip. We hope to be able to illuminate the mechanism(s) which govern the crossover from elastic to plastic regime, the buckling (phase) transitions, and the mechanical fracture in these systems. We use GPU-based speedup to accelerate biomolecular simulations. For example, an a simgle GPU GeForce GTX 480, it takes ~16 days to generate one force-indentation trajectory for the viral shell CCMV (35,000 residues) using the SOP-GPU package and the experimental \(\sim 1 \mu m/s\) force-load.
Forced indentation in silico of the bacteriophage HK97¶
Topology and the arrangement of the secondary structure elements into the overall structure, rather than atomic details, govern large-scale conformational transitions. We utilize the Self Organized Polymer (SOP) model and Langevin simulations, fully implemented on the GPU (SOP-GPU package), to perform dynamic force experiments in silico of the mechanical properties of the bacteriophage HK97 (115,140 residues). This biological assembly is made of 420 copies of the gp5 protein and is formed by 60 icosahedral units, each composed of 7 domains A-G. The capsid outer radius is ~32 nm, and the average wall thickness is ~2.1 nm (head II state; PDB code 2FT1). We probe the mechanical reaction of the bacteriophage HK97 by indenting it with the time-dependent force \(f(t)=r_ft\), where \(r_f=\kappa \nu_f\) is the force-loading rate, and \(\kappa\) and \(\nu_f\) are, respectively, the cantilever spring constant and the tip velocity. We analyze the dependence of the physical properties of HK97 on the rate of change \(r_f\) and geometry of mechanical perturbation (spherical tip of different radii \(R\)). It takes ~34 days of wall-clock time (\(10^9\) steps) to generate a singe indentation trajectory of length 20 ms using \(\nu_f=2.5 \mu m/s\) on the GPU GeForce GTX 480. We observed a whole spectrum of biomechanical reactions from the gradual indentation to buckling, and to fracture. These results agree well with the experimental observations on other virions. These dynamic signatures can be used to provide meaningful interpretation of the force peaks and kinks in the experimental force-indentation curves.
Continuous indentation¶
Buckling transition¶
Structural failure¶
Cowpea Chlorotic Mottle Virus capsid biomechanics from nanomanipulation in silico¶
Using Self-Organized Polymer model implemented in SOP-GPU package we performed computational modeling on sub-second timescales of the indentation nanomechanics of Cowpea Chlorotic Mottle Virus (CCMV) capsid. The simulations show that the capsid’s physical properties are dynamic and local characteristics of the structure, which depend on the magnitude and geometry of mechanical input. Surprisingly, under large deformations the CCMV capsid transitions to the collapsed state without substantial local structural alterations. The enthalpy change in this deformation state \(\Delta H = 11.5 - 12.8\) MJ/mol is mostly due to large-amplitude out-of-plane excitations, which contribute to the capsid bending, and the entropy change \(T\Delta S = 5.1 - 5.8\) MJ/mol is mostly due to coherent in-plane rearrangements of protein chains, which result in the capsid stiffening. Dynamic coupling of these modes defines the extent of elasticity and reversibility of capsid mechanical deformation. This emerging picture illuminates how unique physico-chemical properties of protein nanoshells help define their structure and morphology, and determine their viruses’ biological function.