Currently accepting PhD student applications (email: frederic_chain[at]uml.edu)
Info on PhD program in Applied Biology at UMass Lowell
Info on PhD program in Applied Biology at UMass Lowell
current lab members
|
Phd candidate Environmental metagenomics of invasive mosquitoes: the effects of environmental stress on microbiome diversity and activity *Master's from Brandeis University *Clinical field application specialist for Roche Diagnostics *Aims to develop new approaches for treating infectious diseases Master's student Investigation into the role of Huntingtin protein as a scaffolding protein in the autophagy pathway *B.S. in Biotechnology UML (2020) Magna Cum Laude *Senior Research on Huntingtin protein function using the model Dictyostelium discoideum *Currently a Lab Technician at Lahey Hospital and Medical Center, performing COVID testing of patient samples through RT-PCR master's student Copy Number Variation Mutation Rates in Plasmodium *B.S. at UML (2021) *Research on iron and magnesium dependence of huntingtin deficient Dictyostelium cells *Concentration in Biotechnology BIO UNDERGRAD single-cell RNA-seq analysis of primate brains *Honors college fellowship (2021) *Concentration in Bioinformatics immersive scholar |
Master's student eDNA metabarcoding to monitor aquatic biodiversity during Alewife migration *Senior research on molecular approaches in metabarcoding *Conducted in silico analysis of metabarcoding primer pairs to evaluate the extent of taxonomic diversity and resolution Master's student Tardigrade genomics, immunology, and gene regulation *B.S. at UML (2020) Magna Cum Laude *Senior Thesis on bacterial plate counts from glove boxes in a hospital setting *Phlebotomist *Part-time assistant - laboratory animal care and maintenance *Goal is to pursue PhD immersive scholar 16S and 18S profiling of environmental samples and macroinvertebrate diversity *Immersive Scholar (2021) *Concentration in Biotechnology immersive scholar immersive scholar 16S and 18S profiling of environmental samples and macroinvertebrate diversity *Immersive Scholar (2021) *Majoring in Env. Engineering |
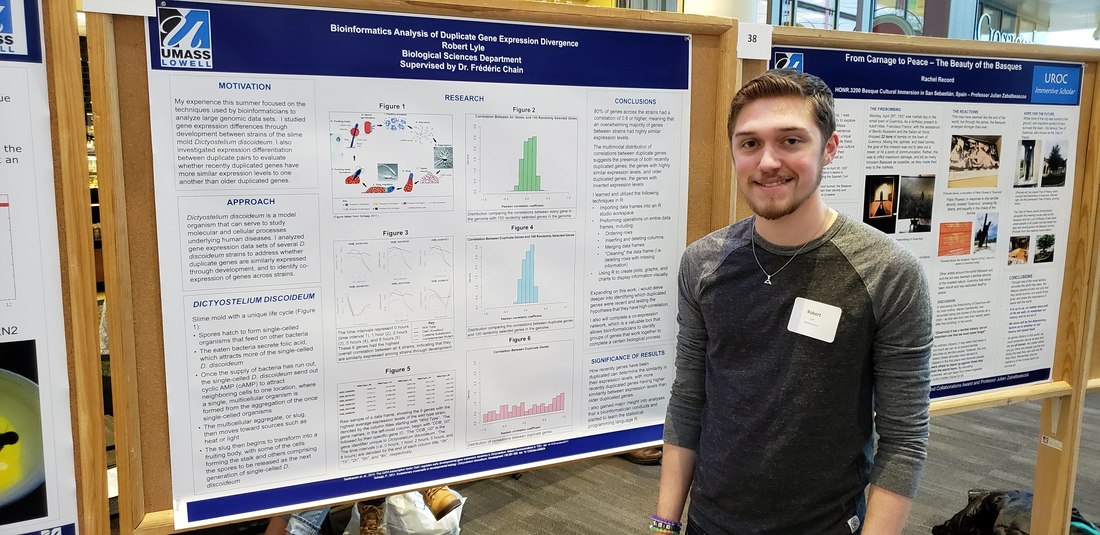
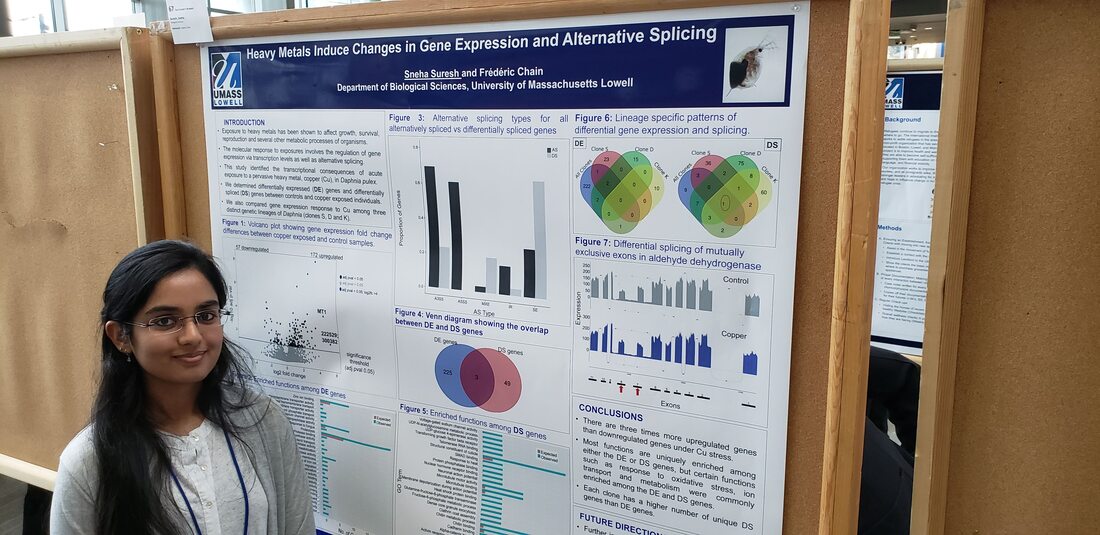
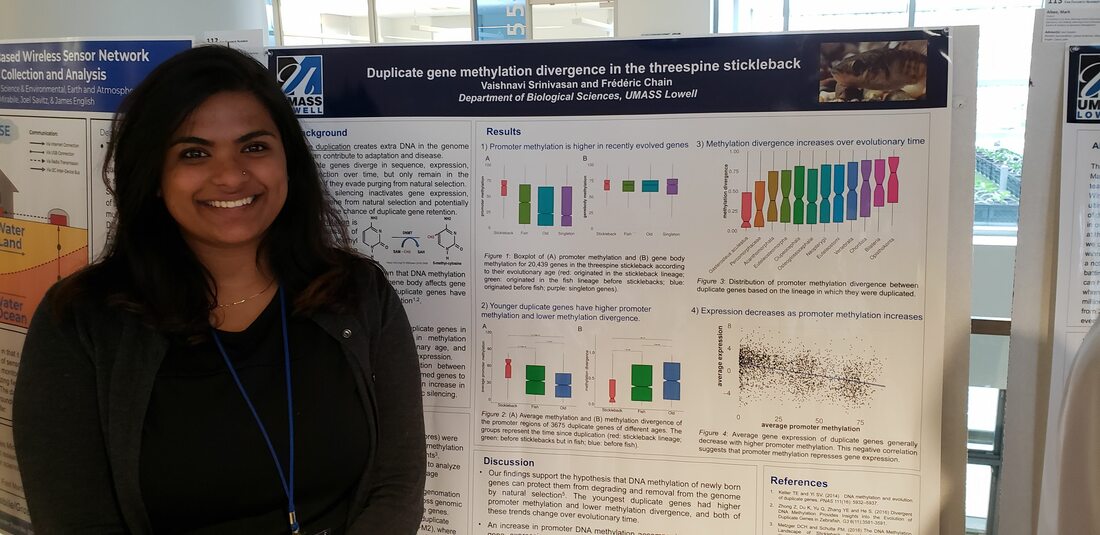
Master's student Duplicate gene expression variation in single-cell RNA-seq *Bsc in Bioinformatics - UML *Programming experience in R, python and C/C++ *Goal is to apply bioinformatics tools to study gene functions Dictyostelium epigenetics, duplicate gene evolution *B.S. in biology from UML (2021) Summa Cum Laude *Senior Research on single cell methylation and expression *Teaching Assistant *Summer Research Assistant * Goal is to develop/apply bioinformatic tools that can aid in biology and health research. bio undergrad Copy Number Variation mutation rates *Co-op at uniQure gene therapy *Senior Research (2021) *Honors college fellowship (2020) *Immersive Scholar (2019) UNdergrad Volunteer eDNA metabarcoding bioinformatic analysis *Honors Student in Biology Principal investigator genome evolution duplicate genes mutation rates eDNA biodiversity monitoring microbial profiling alternative splicing |
lab alumni
2020-2021
|
honors project Huntingtin deficiency in Dictyostelium development & expression *KCS Scholar (2019) *2021 UMass Lowell Trustees’ Key and Chancellor’s Distinguished Academic Achievement Medal *2021 Biology Department Academic Achievement Award (Top GPA) *Medical ophthalmology assistant |
senior thesis Microbial metagenomics & Dictyostelium mutagenesis *UMLSAMP Scholar (2020) *Graduated in Biology with a Minor in Public Health (Cum Laude) *College of Science Dean's List *Intern at the Smithsonian Astrophysical Observatory 2020 *Pharmacy School @ University of Wisconsin Madison |
|
immersive scholar immersive scholar eDNA metabarcoding processing methods *Exercise Science Major (Pre-Med) |
2019-2020
|
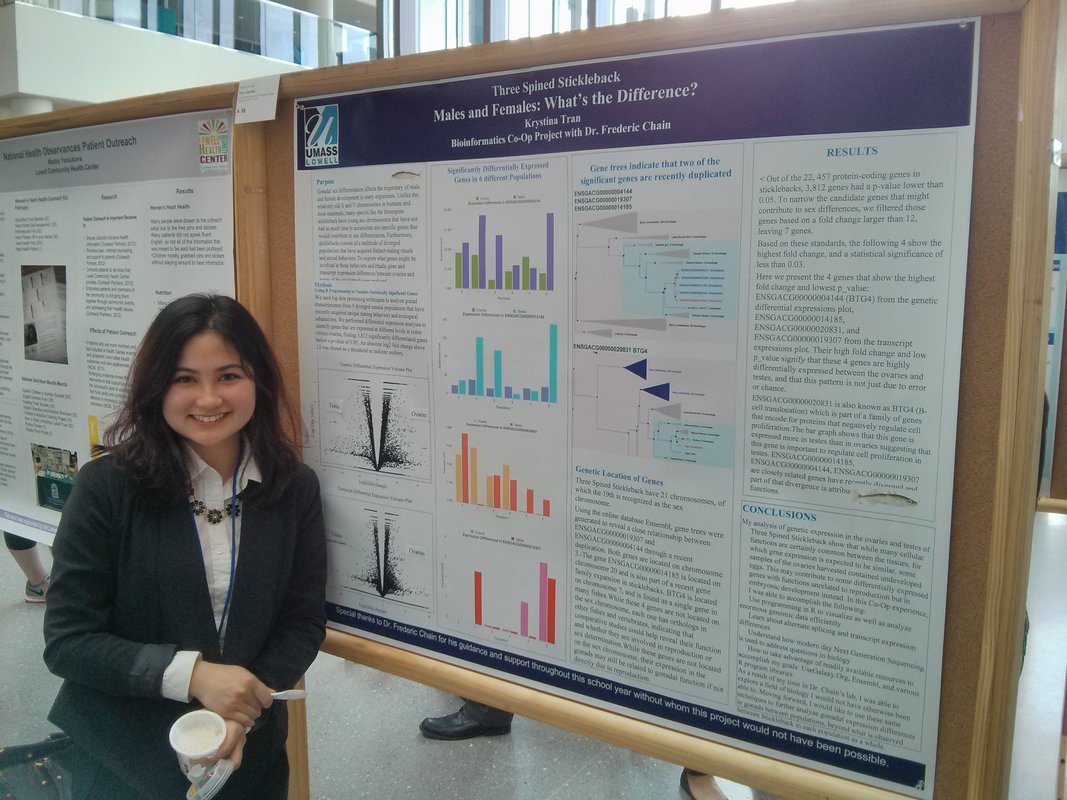
volunteer Differential Gene Splicing *Research Associate @ Cystic Fibrosis Foundation Therapeutics Lab |
|
2018-2019
|
|
|
2017-2018
|
|
|
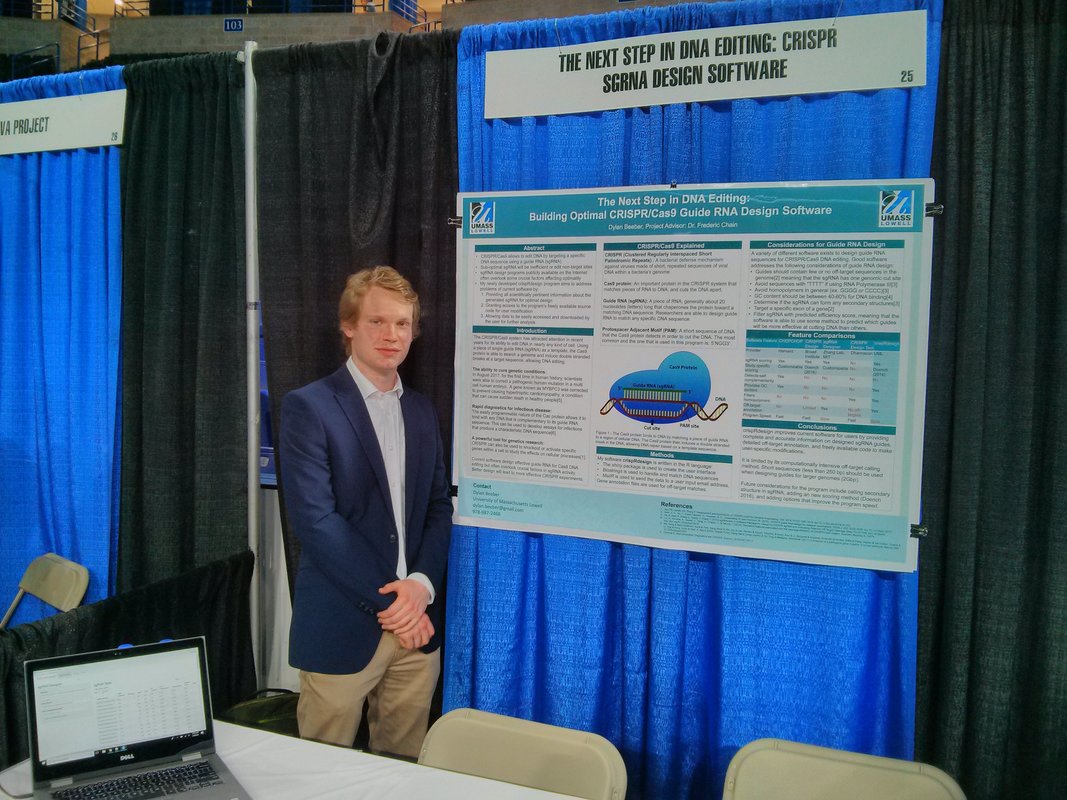
Dylan Beeber Honors thesis Developed an R program to design guide RNA for CRISPR applications Software found here * Hired @ SQZ biotech Max Kalinowski Monitoring urban waterways for pollutants and biodiversity |
Mapping pollutants and biodiversity in canals & DNA Methylation between populations |
Vertical Divider
lab guests
Donghyun Lee (2020-2021) - Structural variation calling using long read technologies
Guy Moore (2018-2019) - Comparing metagenomics analysis and taxonomic assignment pipelines
Louis Astorg (2018) - PhD Candidate at UQAM studying eco-evolutionary dynamics in ionic gradients
Gillian Martin (2017) - PhD Candidate at UQAM studying the impact of dispersal and genetic diversity on metacommunity dynamics
Guy Moore (2018-2019) - Comparing metagenomics analysis and taxonomic assignment pipelines
Louis Astorg (2018) - PhD Candidate at UQAM studying eco-evolutionary dynamics in ionic gradients
Gillian Martin (2017) - PhD Candidate at UQAM studying the impact of dispersal and genetic diversity on metacommunity dynamics
Vertical Divider
some collaborators
Xenopus Genomics
Ben Evans @ McMaster University
Daphnia Genomics and Biodiversity
Melania Cristescu @ McGill University
Teri Crease @ Guelph University
Stickleback Genomics
Philine Feulner @ eawag
Natalie Steinel @ UMass Lowell
Anne Dalziel @ Saint Mary's University
Britta Meyer @ Max Planck Institute for Evolutionary Biology
Erich Bornberg-Bauer @ University of Münster
Christophe Eizaguirre @ Queen Mary University of London
Thorsten Reusch @ GEOMAR Kiel
Jason Goldstein @ Wells National Estuarine Research Reserve
Environmental Monitoring in Lowell
Sheree Pagsuyoin @ UMass Lowell
Grace Chen @ UMass Lowell
Daniel Obrist @ UMass Lowell
Richard Gaschnig @ UMass Lowell
Jane Calvin @ Lowell Parks & Conservation Trust
Sheila Kirschbaum @ UMass Lowell & Tsongas Industrial History Center
Mutation, Stress, and Transcriptomics
Natalie Steinel @ UMass Lowell
Jennifer Fish @ UMass Lowell
Mike Myre @ UMass Lowell
Ben Evans @ McMaster University
Daphnia Genomics and Biodiversity
Melania Cristescu @ McGill University
Teri Crease @ Guelph University
Stickleback Genomics
Philine Feulner @ eawag
Natalie Steinel @ UMass Lowell
Anne Dalziel @ Saint Mary's University
Britta Meyer @ Max Planck Institute for Evolutionary Biology
Erich Bornberg-Bauer @ University of Münster
Christophe Eizaguirre @ Queen Mary University of London
Thorsten Reusch @ GEOMAR Kiel
Jason Goldstein @ Wells National Estuarine Research Reserve
Environmental Monitoring in Lowell
Sheree Pagsuyoin @ UMass Lowell
Grace Chen @ UMass Lowell
Daniel Obrist @ UMass Lowell
Richard Gaschnig @ UMass Lowell
Jane Calvin @ Lowell Parks & Conservation Trust
Sheila Kirschbaum @ UMass Lowell & Tsongas Industrial History Center
Mutation, Stress, and Transcriptomics
Natalie Steinel @ UMass Lowell
Jennifer Fish @ UMass Lowell
Mike Myre @ UMass Lowell