Currently accepting PhD student applications (email: frederic_chain[at]uml.edu)
Info on PhD program in Applied Biology at UMass Lowell
Info on PhD program in Applied Biology at UMass Lowell
genome evolution, ecological adaptation and speciation
Vertical Divider

Genome-wide variation in recently diverged populations can reveal molecular signatures reflecting the processes driving adaptive evolution and speciation. The three-spined stickleback is a useful vertebrate model to investigate genome evolution during population differentiation and ecological adaptation; freshwater colonization has repeatedly occurred since the last glaciation giving rise to recurrent phenotypic diversification and locally-adapted ecotypes.
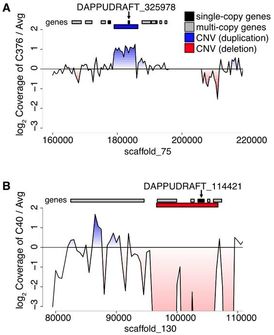
As part of a consortium headed by Manfred Milinski, we carried out comparative & population genomics analyses and immunological transcriptomics to assess the extent of genome-wide differentiation among pairs of lake-river ecotypes. Results indicate that selection is a major contributor to the observed heterogeneous patterns of genomic divergence, and that structural variation substantially contributes to genetic variation. Young genes such as recently duplicated genes are overrepresented among copy-number variations (CNVs), and are enriched with functional roles related to environmental response. These young duplicates provide ample raw genetic material that may be used for adaptation to novel environments.
Current investigations include the emergence of new genes across populations and the role of epigenetics in determining the regulation and evolutionary fate of new genes.
As part of a consortium headed by Manfred Milinski, we carried out comparative & population genomics analyses and immunological transcriptomics to assess the extent of genome-wide differentiation among pairs of lake-river ecotypes. Results indicate that selection is a major contributor to the observed heterogeneous patterns of genomic divergence, and that structural variation substantially contributes to genetic variation. Young genes such as recently duplicated genes are overrepresented among copy-number variations (CNVs), and are enriched with functional roles related to environmental response. These young duplicates provide ample raw genetic material that may be used for adaptation to novel environments.
Current investigations include the emergence of new genes across populations and the role of epigenetics in determining the regulation and evolutionary fate of new genes.